Terminology
- RPKM (Reads Per Kilobase Million) is for single end RNA-seq.
- FPKM (Fragments Per Kilobase Million) is for paired end RNA-seq.
- TPM (Transcripts Per Million)

- Gene-level quantification : count the number of reads that map (i.e. align) to each gene (read count)
- library : In molecular biology, a library is a collection of DNA fragments that is stored and propagated in a population of micro-organisms through the process of molecular cloning.
import pandas as pd
df=pd.read_table('quant.sf',sep='\t')
df['RPK']=df['NumReads']*1000/df['EffectiveLength'] # unit in kilobase
scale_factor=df['RPK'].sum()/1000000
df['TPM']=df['RPK']/scale_factor
For quantifying RNA-seq, Salmon is used. In order to run the Salmon, It needs reference fasta (index) and RNA-seq fastas.
- Reference fasta to indexing : ftp://ftp.ensembl.org/pub/current_fasta/homo_sapiens/cdna/Homo_sapiens.GRCh38.cdna.all.fa.gz
$ wget ftp://ftp.ensembl.org/pub/current_fasta/homo_sapiens/cdna/Homo_sapiens.GRCh38.cdna.all.fa.gz
$ gunzip Homo_sapiens.GRCh38.cdna.all.fa.gz
$ salmon index -t Homo_sapiens.GRCh38.cdna.all.fa -i index_directory_name
- Trim a sample's fasta file
https://www.hadriengourle.com/tutorials/qc/
$ sickle
- Quantify
$ salmon quant -i salmon_index -l ISF -1 sample_R1.fasta -2 sample_R2.fasta -o quant
# the library format option (-l) comes before the read libraries.
$ cat quant.sf
Name Length EffectiveLength TPM NumReads
ENST00000632638.1 552 327.266 10.897145 0.500
ENST00000631824.1 406 185.067 0.000000 0.000
ENST00000632308.1 380 161.138 88.527190 2.000
ENST00000610439.4 381 162.030 0.000000 0.000
ENST00000631882.1 397 176.542 40.401413 1.000
Each subsequent row describes a single quantification record. The columns have the following interpretation.
* Name — This is the name of the target transcript provided in the input transcript database (FASTA file).
* Length — This is the length of the target transcript in nucleotides.
* EffectiveLength — This is the computed effective length of the target transcript. It takes into account all factors being modeled that will effect the probability of sampling fragments from this transcript, including the fragment length distribution and sequence-specific and gc-fragment bias (if they are being modeled).
: l^\hat_i (Effective length) = l_i (transcript length) - u_FLD (mean fragment length) + 1
* TPM — This is salmon’s estimate of the relative abundance of this transcript in units of Transcripts Per Million (TPM). TPM is the recommended relative abundance measure to use for downstream analysis.
* NumReads — This is salmon’s estimate of the number of reads mapping to each transcript that was quantified. It is an “estimate” insofar as it is the expected number of reads that have originated from each transcript given the structure of the uniquely mapping and multi-mapping reads and the relative abundance estimates for each transcript.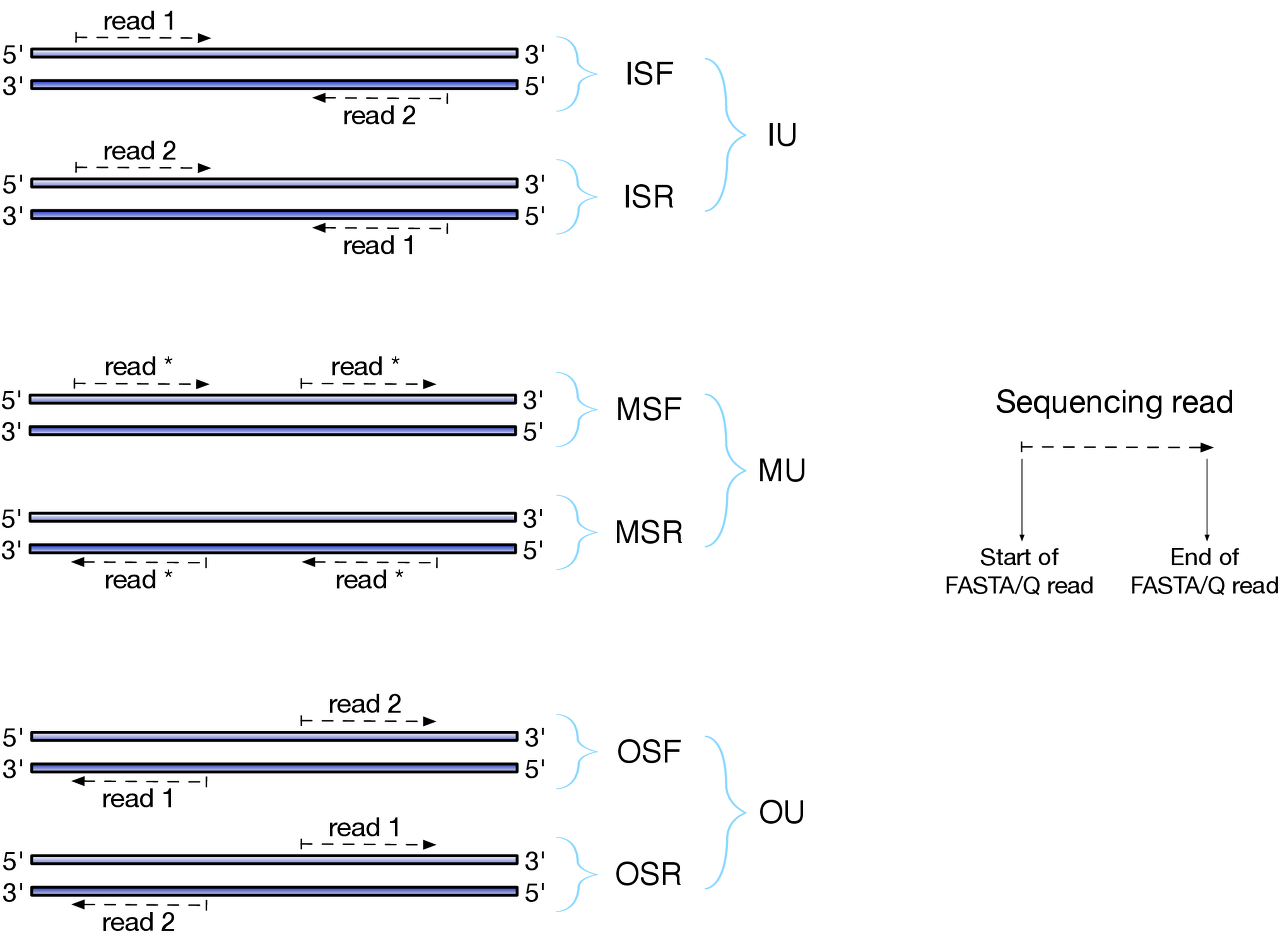
Map from transcript ID to gene
library(tximport)
library(readr)
tx2gene <- read_csv(file.path("/path/salmon_index","tx2gene.csv"))
txi <- tximport("quant.sf",type="salmon",tx2gene = tx2gene, countsFromAbundance=c("lengthScaledTPM"))
write.table(txi,"quant2.sf",sep="\t",row.names=TRUE,na="NA") "abundance" "counts" "length"
"5_8S_rRNA" 0 0 5.66531
"5S_rRNA" 9.90597 5.13146602513917 15.3415770868476
"7SK" 0 0 70.9598545454545
"A1BG" 1.4734829 28.272575130701 568.258347199007
"A1BG-AS1" 1.491531016 50.0242444818419 993.284838677479
"A1CF" 0.0268588 8.14734841349264 8983.7
"A2M" 0.2695483 7.00000363356594 769.10781708473
"A2M-AS1" 0.106058 6.99999917610522 1954.7
"A2ML1" 0.255674 24.5561890720357 2844.46084398101- abundance : sum(TPM)
- counts : sum(NumReads)
Fold changes for finding differentially expressed genes
: FC = treatment / control
: 방향성을 보기 위해 log값을 구함, log2(FC).
https://bioconductor.riken.jp/packages/3.4/bioc/vignettes/tximport/inst/doc/tximport.html
https://blog.naver.com/guhwang/222136013429
https://blog.naver.com/naturelove87/221454634945
https://www.reneshbedre.com/blog/expression_units.html
https://www.biostars.org/p/342756/
What is the correct way to understand a fold change value of a gene or protein?
- A foldchange describes the difference of two values
(eg. difference of expression in gene/protein A between healthy and diseased case) - Biostatistical porgrams/packages calculate it via: "Log(FC)" = mean(log2(Group1)) - mean(log2(Group2))
- log2 fold changes are used/plotted in graphs as those are nicer to show because they center around 0, giving reductions a negative value and increments a positive value
- log2 fold change values (eg 1 or 2 or 3) can be converted to fold changes by taking 2^1 or 2^2 or 2^3 = 1 or 4 or 8
- To convert the fold change into change in % or anything that is actually tangible/understandable in "real life terms" ... need answers here! (= actual question I want to ask)
https://www.reddit.com/r/labrats/comments/7odtki/dumb_question_about_logfc/
... if logFC = -0.5, then FC = 2^(-0.5), or 0.7071, which means about 70% of the baseline, not 50%... 50% reduction in expression would be a logFC of -1. ...
Main step for RNA-seq
1) Prepare a sequencing library
s1. Isolate RNA
s2. Break the RNA into small fragments
s3. Convert the RNA fragments into double stranded DNA : Double stranded DNA is more stable than RNA and can be easily amplified and modified.
s4. Add sequence adaptors : 1) Allow the sequening machine to recognize the fragments 2) Allow you to sequnce different samples at the same time, since different samples can use different adaptors
s5. PCR amplify
s6. QC : 1) Verify library concentration 2) Verify library fragment lenths
2) Sequence
s1. Plot the data, PCA for logFC
s2. Identify differentially expressed genes between the "normal" and "mutant" samples.

3) Data analysis
https://www.researchgate.net/post/How_to_calculate_the_log2_fold_change
Fold change is a measure describing how much a quantity changes going from an initial to a final value. For example, an initial value of 30 and a final value of 60 corresponds to a fold change of 2 (or equivalently, a change to 2 times), or in common terms, a one-fold increase. Fold change is calculated simply as the ratio of the difference between final value and the initial value over the original value. Thus, if the initial value is A and final value is B, the fold change is (B - A)/A or equivalently B/A - 1. As another example, a change from 80 to 20 would be a fold change of -0.75, while a change from 20 to 80 would be a fold change of 3 (a change of 3 to 4 times the original).
Fold change is often used in analysis of gene expression data in micro array and RNA-Seq experiments, for measuring change in the expression level of a gene.[6] A disadvantage to and serious risk of using fold change in this setting is that it is biased [7] and may miss deferentially expressed genes with large differences (B-A) but small ratios (A/B), leading to a high miss rate at high intensities.
Let's say there are 50 read counts in control and 100 read counts in treatment for gene A. This means gene A is expressing twice in treatment as compared to control (100 divided by 50 =2) or fold change is 2. This works well for over expressed genes as the number directly corresponds to how many times a gene is overexpressed. But when it is other way round (i.e, treatment 50, control 100), the value of fold change will be 0.5 (all under expressed genes will have values between 0 to 1, while over expressed genes will have values from 1 to infinity). To make this leveled, we use log2 for expressing the fold change. I.e, log2 of 2 is 1 and log2 of 0.5 is -1.
Reference
- https://www.biostars.org/p/342756/
- https://www.reddit.com/r/labrats/comments/7odtki/dumb_question_about_logfc/
- https://en.wikipedia.org/wiki/Library_(biology)
- https://salmon.readthedocs.io/en/latest/index.html
- https://hbctraining.github.io/Intro-to-rnaseq-hpc-orchestra/lessons/09_salmon.html
'Biology' 카테고리의 다른 글
| Gene family (0) | 2021.06.01 |
|---|---|
| DESeq2 - Library normalization (0) | 2021.05.18 |
| Heritability (0) | 2021.05.17 |
| mRNA Vaccine (0) | 2021.05.08 |
| 암치료에 대한 생각 (0) | 2021.04.26 |



댓글