
Phasing을 위한 tool이 많이 있지만, shapeit4에 대해서 정리해 본다.
공식 홈페이지(https://odelaneau.github.io/shapeit4/)가 있으며, source code는 github(https://github.com/odelaneau/shapeit4)에 있다.
Shapeit4의 설치를 위해서는 몇 가지 library가 필요하다.
- HTSlib: A great C library for reading/writing high-throuhput sequencing data.
- BOOST: A free peer-reviewd portable C++ source libraries. SHAPEIT4 uses two specific BOOST libraries: iostreams and program_options.
# git 2.x
$ sudo rpm -Uvh http://opensource.wandisco.com/centos/7/git/x86_64/wandisco-git-release-7-2.noarch.rpm
$ sudo yum install git
# gcc 7.x
$ sudo yum install centos-release-scl
$ sudo yum install devtoolset-7
$ scl enable devtoolset-7 bash
# zstd
$ git clone https://github.com/Microsoft/vcpkg.git
$ cd vcpkg
$ ./bootstrap-vcpkg.sh
$ ./vcpkg integrate install
$ ./vcpkg install zstd
# install mlocate
$ sudo yum -y install mlocate
$ sudo updatedb
# export path for 'pyconfig.h'
$ locate pyconfig.h
$ export CPLUS_INCLUDE_PATH="$CPLUS_INCLUDE_PATH:/home/user/python3/"
$ sudo yum -y install libcurl-devel openssl-devel bzip2-devel xz-devel libffi-devel ncurses-devel libevent-devel vcftools
# HTSlib
$ wget https://github.com/samtools/htslib/releases/download/1.10.2/htslib-1.10.2.tar.bz2
$ tar -xvf htslib-1.10.2.tar.bz2
$ cd htslib-1.10.2
$ ./configure
$ make && sudo make install
# BOOST
$ wget https://dl.bintray.com/boostorg/release/1.73.0/source/boost_1_73_0.tar.bz2
$ tar -xvf boost_1_73_0.tar.bz2
$ cd boost_1_73_0
$ ./bootstrap.sh
$ ./b2 install
makefile에 path를 잘 설정해야 프로그램의 실행이 문제없이 진행된다.
$ git clone https://github.com/odelaneau/shapeit4.git
$ cd shapeit4
$ locate libboost_program_options.a libboost_iostreams.a libhts.a
$ emacs makefile
HTSLIB_INC (line 5): path to the HTSlib header files
HTSLIB_LIB (line 6): path to the static HTSlib library (file libhts.a)
BOOST_INC (line 9): path to the BOOST header files (often /usr/include)
BOOST_LIB_IO (line 10): path to the static BOOST iostreams library (file libboost_iostreams.a)
BOOST_LIB_PO (line 11): path to the static BOOST program_options library (file libboost_program_options.a)
Add libraries (line 32): DYN_LIBS=-lz -lbz2 -lm -lpthread -llzma -lcurl -lssl -lcrypto
$ make
위 설치과정이 번거롭거나 어려움이 있으면, docker를 사용해도 된다.
$ sudo yum -y install docker docker-registry
$ sudo systemctl enable docker.service
$ sudo systemctl start docker.service
$ sudo systemctl status docker.service
$ sudo docker pull lifebitai/shapeit4
$ sudo docker images
$ sudo docker run -i -t docker.io/lifebitai/shapeit4 /bin/bash
$ sudo docker ps -a
$ sudo docker start container-ID
$ sudo docker attach container-ID
REFERENCE DATA
기본적으로 UKBiobank data를 받기 위해서는 key가 있어야 한다. 그러기 위해서는 연구자로 등록하고 research에 대해서 application을 제출해야한다.
key가 준비되어 있다면, 프로그램인 ukbgene을 다운받는다. ukbgene은 LINUX에서 작동한다.
$ wget -nd biobank.ctsu.ox.ac.uk/crystal/util/ukbgene
$ chmod 755 ukbgene
$ ukbgene hap -ak12345.key
Usage: ukbgene datatype [flags]
-a authentication file (application_id + 24-char key)
-c chromosome (1-26, X, Y, XY or MT)
-d name of output datafile
-h show this usage message then exit
-i show program version information only then exit
-m fetch mapping/family file associated with datatype
-v verbose mode on
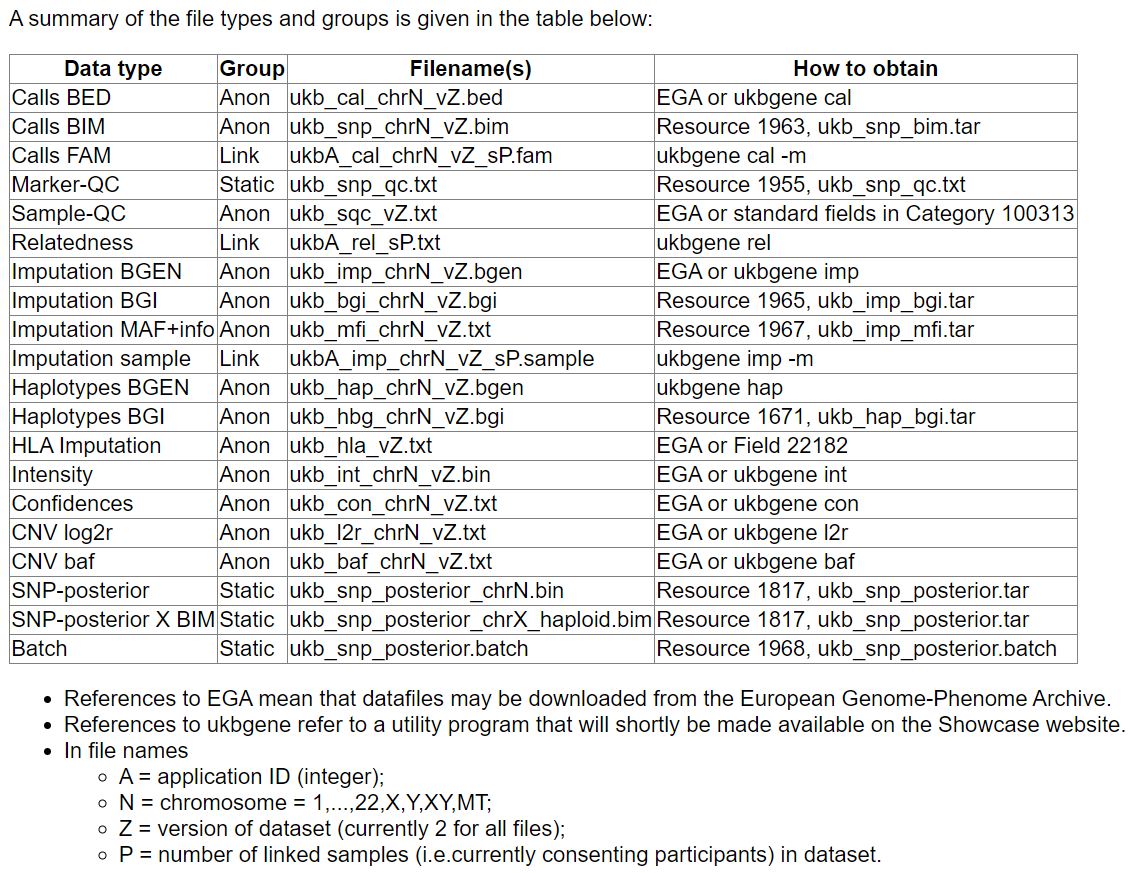
Imputation과 Haplotype에 대한 정보는 아래 링크에 있다.
https://biobank.ctsu.ox.ac.uk/crystal/label.cgi?id=100319
다양한 corhort의 데이터의 사이트 주소를 알려준다.
Data : ftp://ftp.ncbi.nlm.nih.gov/hapmap/phasing/2009-02_phaseIII/HapMap3_r2/

Data : http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/
(Fully phased 2504 individuals)
Reference
- https://odelaneau.github.io/shapeit4/
- https://github.com/odelaneau/shapeit4
- http://biobank.ctsu.ox.ac.uk/crystal/crystal/docs/ukbgene_instruct.html
- www.haplotype-reference-consortium.org/
- https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html
- https://www.internationalgenome.org/category/imputation/
- http://csg.sph.umich.edu/abecasis/MaCH/download/
'Tools' 카테고리의 다른 글
| checkVCF (0) | 2020.09.15 |
|---|---|
| Windows Terminal (0) | 2020.08.17 |
| Slurm - Workload manager (0) | 2020.08.15 |
| GATK (0) | 2020.08.07 |
| LDpop (0) | 2020.07.12 |




댓글