
SCSA: cell type annotation for single-cell RNA-seq data
- Paper : https://www.frontiersin.org/articles/10.3389/fgene.2020.00490/full
- Github : https://github.com/bioinfo-ibms-pumc/SCSA
DB format
: two-column data table, [ 'cell-type', 'marker-gene']
from CellMarker
| Cell marker | Cell type | Tissue | |
| HUMAN | 11464 | 456 | 158 |
| MOUSE | 7855 | 385 | 80 |
and CancerSEA, https://github.com/camlab-bioml/cancersea
: 14 functional states from 25 human cancer type
Input
: DEGs clusters matrix from the clustering output of CellRanger or Seurat.
Filteration
: log2-based fold-change (LFC) value and P-value (LFC ≥1, P ≤ 0.05).
GO Enrichment analysis
: uses Fisher's exact test and Benjamini-Hochberg adjustment.
(using DEGs of the selected cluster as foreground values and DEGs in other clusters as background values)
Limitation
- The result depend on the quantity of marker genes in these cell marker databases.
> use DB with user-defined marker data.
- The accuracy of cell annotation is heavily relied on the clustering algorithms.
> supervised clustering may be more appropriate for cell type classification.
Examples
# To annotate a human scRNA-seq sets generated by CellRanger, use the following code
$ python3 SCSA.py -d whole.db -i cellranger_pbmc_3k.csv -k All -g Human -p 0.01 -f 1.5 -m txt -o sc.txt
# To annotate a human scRNA-seq sets generated by 'FindAllMarkers' function of Seurat(Butler, A., et al. Nature Biotechnology. 2018) with ensemblIDs, use the following code
$ python3 SCSA.py -d whole.db -s seurat -i seurat_GSE72056.csv -k All -E -g Human -p 0.01 -f 1.5
# To annotate a human scRNA-seq sets generated by Scanpy, use the following code
$ python3 SCSA.py -d whole.db -i scanpy_pbmc_3k.csv -s scanpy -E -f1.5 -p 0.01 -o result -m txt
# To annotate a human scRNA-seq sets generated by Scran, use the following code
$ python3 SCSA.py -d whole.db -s scran -i scran_pbmc_3k.csv -k All -g Human -p 0.05 -f 1.1 -b
# To annotate a human scRNA-seq sets generated by 'FindAllMarkers' function of Seurat(Butler, A., et al. Nature Biotechnology. 2018) with both user-defined database and CellMarker database, use the following code
$ python3 SCSA.py -d whole.db -i seurat_GSE72056.csv -s seurat -E -f1.5 -p 0.01 -o result -m txt -M user.table
# To annotate a human scRNA-seq sets generated by CellRanger only with user-defined database without any detail print, use the following code
$ python3 SCSA.py -d whole.db -i cellranger_pbmc_3k.csv -f1.5 -p 0.01 -m txt -M user.table -N -b
# To annotate cluster1 of mouse scRNA-seq sets and To annotate cluster1 of mouse scRNA-seq sets generated by CellRanger, use the following code
$ python3 SCSA.py -d whole.db -s seurat -i seurat_mouse.csv -k All -E -g Mouse -p 0.01 -f 1 -m txt -o testout -c 1
# To list tissue names in the SCSA annotation database, use the following code
$ python3 SCSA.py -i none -d whole.db -l
Reference
- http://geneontology.org/docs/ontology-documentation/
- CellMarker
: http://bio-bigdata.hrbmu.edu.cn/CellMarker/index.jsp
: https://doi.org/10.1093/nar/gky900
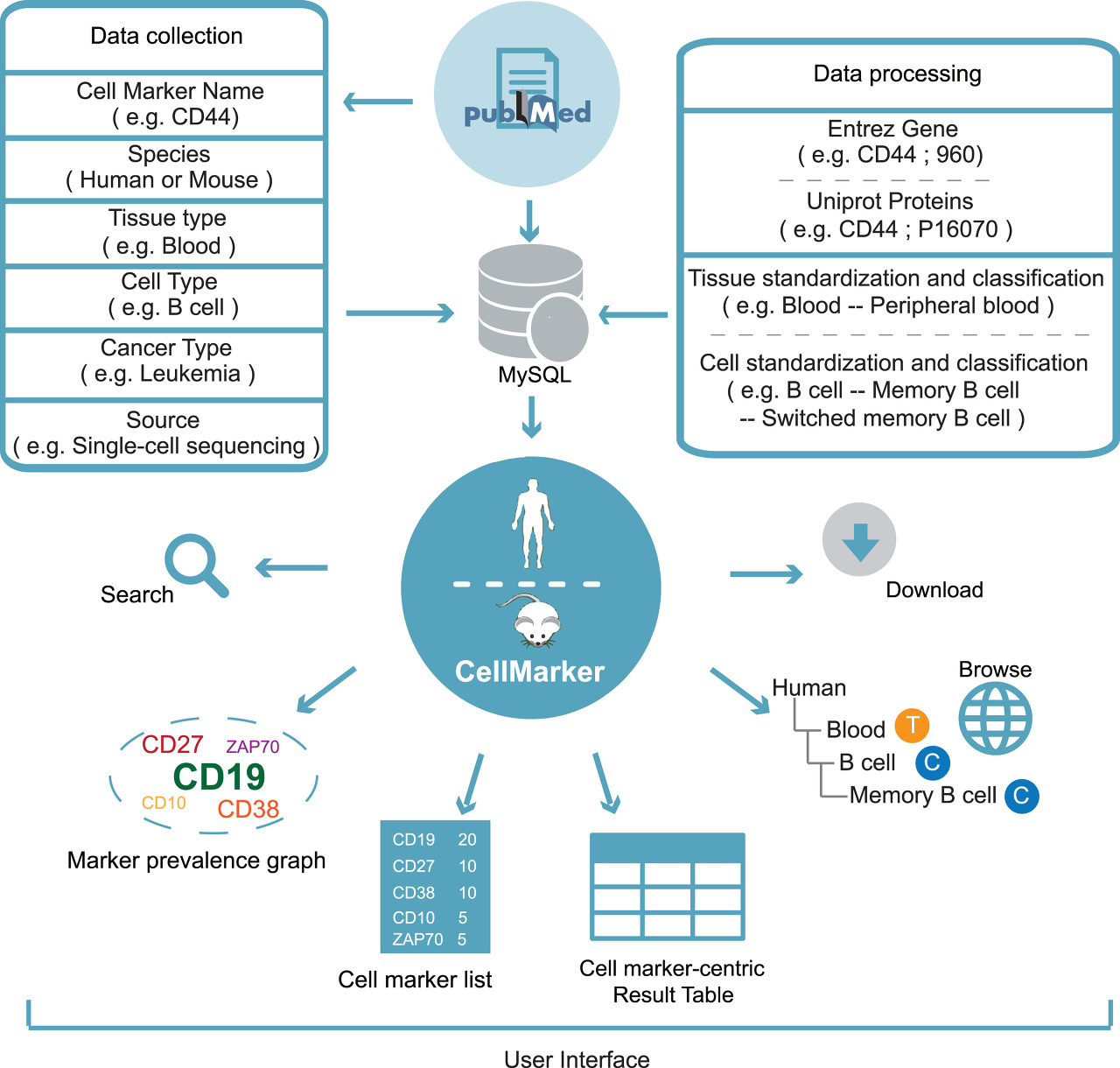
Introduction for CellMarker
CellMarker database aims to provide a comprehensive and accurate resource of cell markers for various cell types in tissues of human and mouse.
Overview for CellMarker
By manually curating over 100,000 published papers, 4,124 entries including the cell marker information, tissue type, cell type, cancer information and source, were recorded. At last, 13,605 cell markers of 467cell types in 158 human tissues/sub-tissues and 9,148 cell makers of 389 cell types in 81 mouse tissues/sub-tissues were collected and deposited in CellMarker.
- CancerSEA
: https://github.com/camlab-bioml/cancersea
: https://doi.org/10.1093/nar/gky939
Introduction for CancerSEA
ScRNA-seq provides an unprecedented opportunity to explore the functional heterogeneity of cancer cells. CancerSEA is the first dedicated database that aims to comprehensively decode distinct functional states of cancer cells at single-cell resolution.
Functions of CancerSEA
1. Providing a cancer single-cell functional state atlas, involving 14 functional states of 41,900 cancer single cells from 25 cancer types.
2. Querying which functional states the gene (including PCG and lncRNA) or gene list of interest is related to across different cancer types.
3. Providing PCG/lncRNA repertoires that are highly related to functional states at single-cell resolution.
Functional states
- Angiogenesis
- Apoptosis
- Cell Cycle
- Differentiation
- DNA damage
- DNA repair
- EMT
- Hypoxia
- Inflammation
- Invasion
- Metastasis
- Proliferation
- Quiescence
- Stemness
Statistic
- No. cancer type 25
- No. cancer single-cells 41900
- No. single-cell datasets 72
- No. cell groups 280
- No. PCGs 18895
- No. lncRNAs 15571

- CSEA-DB
: https://bioinfo.uth.edu/CSEADB/
: https://doi.org/10.1093/nar/gkaa1064
- Human Cell Landscape, http://bis.zju.edu.cn/HCL/contact.html
- Human Cell Atlas, https://www.humancellatlas.org/
- Expression Atlas, https://www.ebi.ac.uk/gxa/download.html
hg38) Gene model tables: ncbiRefSeq, refGene, ensGene, knownGene
http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/
wget http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/hg38.ensGene.gtf.gz
wget http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/hg38.knownGene.gtf.gz
wget http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/hg38.ncbiRefSeq.gtf.gz
wget http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/hg38.refGene.gtf.gz
'Tools' 카테고리의 다른 글
| Marp - Markdown to PPT (0) | 2021.10.13 |
|---|---|
| GSEA (0) | 2021.10.01 |
| Single Cell analysis tools (0) | 2021.08.13 |
| Clustering - Fuzzy (0) | 2021.06.18 |
| VCF reheader (0) | 2021.06.17 |



댓글