보통 우리가 다루는 RNA 데이터는 한 시점에 채취한 혈액이나 tissue에서 얻은 것이다. 즉, 시간에 따라 cell이 어떻게 변화하는지 알 수 없는 정적인 데이터이다.
이러한 정적인 데이터로부터, unspliced mRNA 와 spliced mRNA 의 비율을 통해 cell fate, cell lineage, dynamic pathway 또는 cellular differentiation 를 추정하는 방법이 있다. (RNA velocity of single cells, 2018)
좀 더 robust한 방법으로 likelihood-based dynamic model이 있다. (Generalizing RNA velocity to transient cell states through dynamical modeling, 2020)

Trajectory
- ( Unspliced > spliced ) → ( Positive velocity = Upregulated cell = Highly expressed ) = Start point
- ( Unspliced < spliced ) → ( Negative velocity = Downregulated cell = Lowly expressed ) = End point
- Direction : Downregulated cell → Upregulated cell

Usage (time dependent developing object)
- Stem cell
- 세포 형성에 참여하는 유전자를 찾을 때.
- 세포가 분화하는 동안 과연하는 유전자.
* Cellular Differentiation
: the process of a cell changing from one cell type to another, typically from a less specialized type (stem cell) to a more specialized type (organ/tissue specific cell, eg, colonocyte).
* Gene regulatory networks
: a set of genes, or parts of genes, that interact with each other to control a specific cell function. Gene regulatory networks are important in development, differentiation and responding to environmental cues.
( https://www.nature.com/subjects/gene-regulatory-networks )
* Pseudotime
: the ordering induced by trajectory path.

d. 대각선 라인은 steady state. 파란색 곡선은 발현량의 감소, 빨간색 곡선은 발현량의 증가.

a. PCA를 통한 clustering
d. 발현이 일어나는 순서 (PCA)
h. 발현이 일어나는 순서 (t-SNE)
Velocyto
: The steady-state model estimates volocities as the deviation of the observed ratio of unspliced to spliced mRNA from an inferred steady-state ratio.
: The steady-state ratio is approximated with a linear regression on cells found in the lower and upper quantiles where they are expected to have reached steady-state expression levels.
- Assumption : 1) a common splicing rate across genes 2) the presence of at least partial pbservation of the steady-state expression levels in the sampled dat.
- https://velocyto.org/velocyto.py/index.html
- https://github.com/velocyto-team/velocyto.R
[ Python ]
$ pip install numpy scipy cython numba matplotlib scikit-learn h5py click
$ pip install velocytoimport velocyto as vcy[ R ]
$ sudo apt-get install libboost-devlibrary(devtools)
install_github("velocyto-team/velocyto.R")
scVelo
: The likelihood-based dynamical model generalized velocity estimation to transient systems.
: The kinetics are explained with a deterministic and fully decoupled system of linear differential equations with constant kinetic rate parameters.
- Assumption : constant rates of splicing and degradation across cells for each gene.
- https://scvelo.readthedocs.io/
- https://scvelo.readthedocs.io/VelocityBasics/
[ Python ]
$ pip install adata, scanpy
$ pip install numpy, scipy, pandas, scikit-learn, matplotlib
$ pip install python-igraph louvain pybind11 hnswlib
$ pip install scveloimport scvelo as scv
scv.settings.verbosity = 3 # show errors(0), warnings(1), info(2), hints(3)
scv.settings.presenter_view = True # set max width size for presenter view
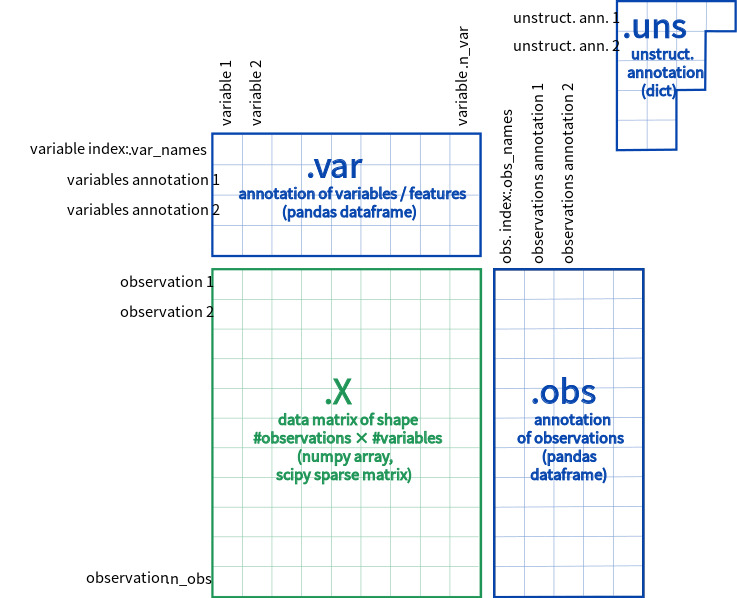
scv.set_figure_params('scvelo') # for beautified visualization- Input : Annotated Data
Reference
- RNA velocity of single cells (2018), https://doi.org/10.1038/s41586-018-0414-6
- Generalizing RNA velocity to transient cell states through dynamical modeling (2020), https://doi.org/10.1038/s41587-020-0591-3
- RNA velocity - current challenges and future perspectives (2021), https://doi.org/10.15252/msb.202110282
- anndata-Annotated Data, https://anndata.readthedocs.io/en/latest/
- RNA splicing and Spliceosome, https://youtu.be/53v2u9ukCt0
- VeloViz, https://github.com/JEFworks-Lab/veloviz
- VeloViz paper, https://doi.org/10.1093/bioinformatics/btab653
'Study' 카테고리의 다른 글
| Standard population (0) | 2022.12.27 |
|---|---|
| Signature matrix (0) | 2021.12.29 |
| scRNA-seq analysis (0) | 2021.08.19 |
| ICGC database (0) | 2021.06.24 |
| Nanopore (0) | 2021.06.22 |


댓글